It has been a while since I entered POSTECH as a freshman in 1997. Admittedly, things have changed quite a bit over the years, and my personal experience is bound to be limited. Still, I am grateful to have a chance to discuss my experience and research topics at POSTECH.
I was fortunate enough to start my graduate studies at the California Institute of Technology in the United States. California Institute of Technology, also known as CALTECH, is a small school that focuses on science and engineering and was also a foundational model for POSTECH when it was established. Indeed, CALTECH has a small student body with about 200 new students enrolled per year and a relatively large body of faculty members. Throughout the campus, interesting science insignia can be found, like Gene Pool and orbital-shaped lamps.
When I started as a fresh graduate student at CALTECH, I had little idea of what I should study as a graduate student. Fortunately for me, there was a “rotation” period for the first year of graduate studies, where all the graduate students would “rotate” through different labs, typically once per quarter (an 11-week period). For the first two semesters, I did a rotation in the neuroscience labs. However, I was strongly drawn to a new research topic “DNA computing,” which was taught by a new faculty member who just started at CALTECH. Later, I had the honor to work with Dr. Erik Winfree as my thesis advisor. My advisor finished his graduate studies at CALTECH, initially working on neural networks in the computer science field but later switched topics to molecular computation mainly using DNA molecules. His thesis advisor was none other than Dr. John Hopfield, who won the Nobel Prize in Physics last year for his foundational studies on the physics of neural networks used for artificial intelligence.
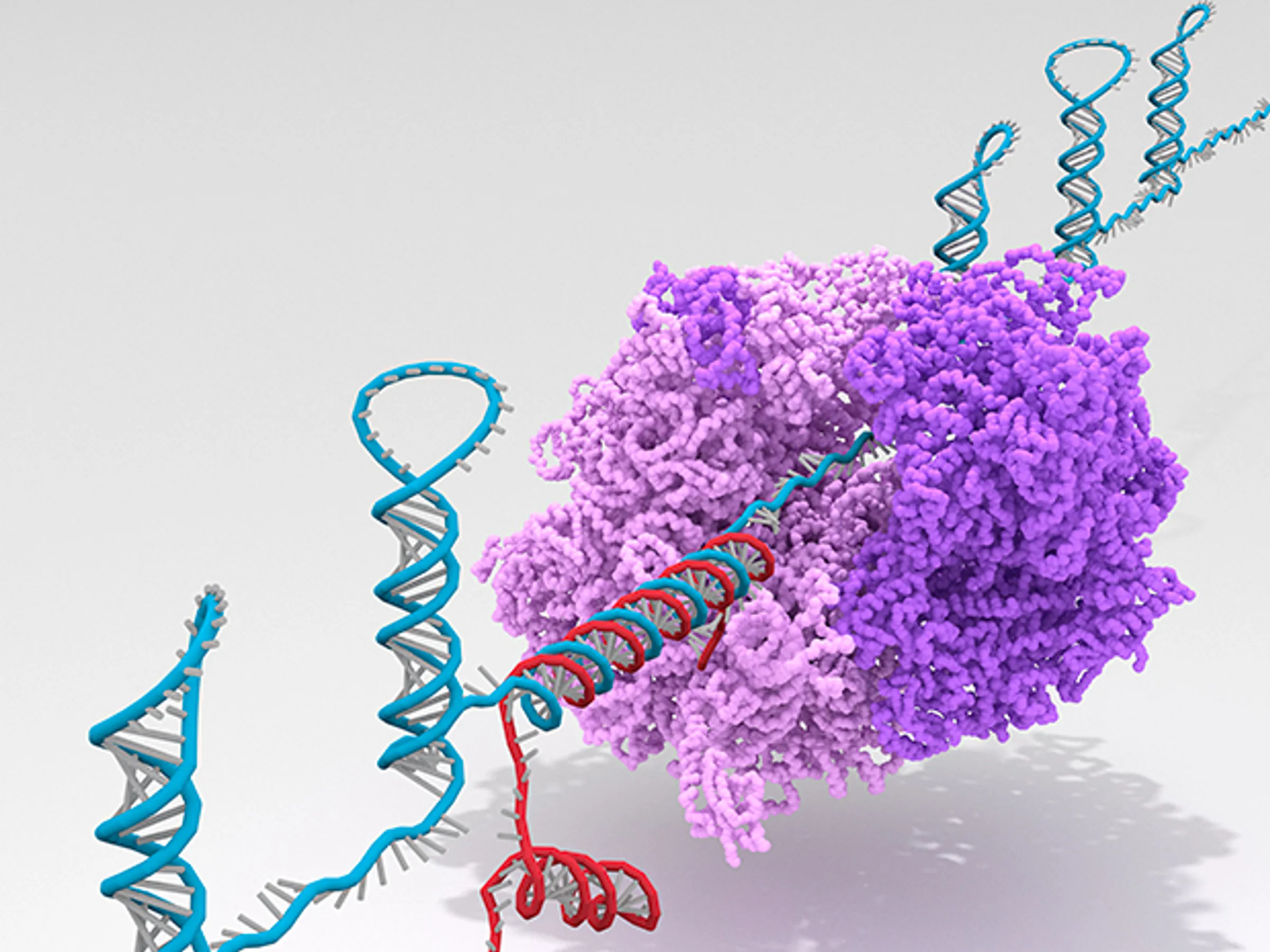
For “DNA Computing,” DNA, rather than as a genetic material for biological organisms, plays a role of information-encoding polymer substrate, where interesting computational tasks can be encoded and operated. It can be further generalized to “Molecular Computing” where biological molecules such as DNA, RNA, and proteins are used for computational tasks in cells. This area is closely related to “Synthetic Biology,” where engineering approaches are used to program biological cells to certain specifications. Coincidentally, the first two seminal papers on “Synthetic Biology” were published back in 2000, although little did I know that I would work on that area of research at that time.
My research involves topics related to “Molecular Computing” and “Synthetic Biology.” As a graduate student and a postdoctoral scholar, my earlier works focused more on “Molecular Computing.” During that time, I developed de–novo-designed transcriptional switches and demonstrated several synthetic programmable transcriptional circuits in test tubes including bistable memory circuits, a fold-change detector, and oscillators interconnected to DNA nanodevices and encapsulated in cell-like volumes.
Later, I shifted my focus towards engineering bacterial cells, developing rationally-designed RNA computing systems as genetically encodable sensors and controllers. These “ribocomputing systems” were built on a recent breakthrough of a nucleic-acid-based gene expression regulator, a “toehold switch,” that provides a library of programmable, orthogonal, and high-performance parts for synthetic biology.
A recent work just published in my group builds on these synthetic RNA devices to generate a new computational framework termed “Synthetic Translation Coupling Element (SynTCE).” In nature, the bacterial genome is replete with “polycistronic operons,” where multiple genes are coordinately expressed to form complexes and perform specific functions. To capitalize on this property, we combined the previously reported ribocomputing system with a newly designed synthetic coupling device that can seamlessly integrate multiple output proteins with programmed stoichiometric balances. The capability to process multiple inputs and multiple outputs within a single RNA transcript is unprecedented. Thus, we believe this synthetic biological platform could be useful for improved functionality in a number of synthetic biological devices.

Together with SynTCE, we also recently published examples on other synthetic platforms that utilize the CRISPR/Cas system and aptamers together with RNA devices. Overall, we are broadly interested in developing synthetic biomolecular systems with precisely prescribed dynamical behavior. By interfacing with biological molecular systems through sensing a biological signal (e.g., mRNA, protein, small molecules) and producing a readout observable by a human user or actuating a biological response, these synthetic molecular devices can provide powerful experimental tools and technological platforms with applications in biosensing, imaging, and potentially in diagnostics and therapeutics.
POSTECH is a unique place where all undergraduate students can be actively engaged in research projects together with excellent faculty members and researchers. I would encourage POSTECH students to take this opportunity to work on research programs at POSTECH and also apply for exchange programs during their undergraduate period. These research experiences would provide great opportunities to learn something valuable from the experience.


